From DNA to Protein: Understanding the Central Dogma of Molecular Biology
Each human cell consists of double-stranded DNA. A single strand of DNA bears specific sequences of four nucleotide bases, namely, adenine (A), thymine (T), cytosine (C), and guanine (G). These nucleotide bases remain in a complementary pairing such that A pairs with T and C pairs with G. Specific sequences of these nucleotide bases form genes. A DNA strand comprises millions of nucleotide bases in various combinations to form genes that encode for a specific protein.
What is the Central Dogma?
A Central Dogma is an explanation of the flow of Genetic information in a cell, including the replication of the DNA, the transcription of the RNA, and the translation of the RNA to create the Proteins. This concept was first proposed by Francis Crick in 1958.
The Central Dogma can be summarised in three main processes:
- Replication: The process by which DNA makes a copy of itself during cell division
- Transcription: The process by which the information in a strand of DNA is copied into a new molecule of messenger RNA (mRNA).
- Translation: The process by which the sequence of the mRNA is used to synthesize a specific proteins. Proteins are formed using the genetic code of the DNA.
Replication
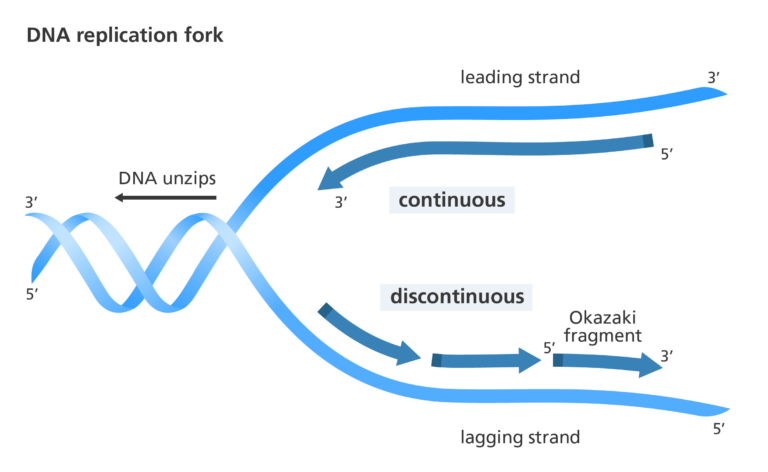
Each time a cell divides, each of its double strands of DNA splits into two single strands. Each of these single strands acts as a template for a new strand of complementary DNA. As a result, each new cell has its own complete genome. This process is known as DNA replication. Replication is controlled by the Watson-Crick pairing of the bases in the template strand with incoming deoxynucleoside triphosphates, and is directed by DNA polymerase enzymes. It is a complex process, particularly in eukaryotes, involving an array of enzymes. A simplified version of bacterial DNA replication is described.
DNA biosynthesis proceeds in the 5'- to 3'-direction. This makes it impossible for DNA polymerases to synthesize both strands simultaneously. A portion of the double helix must first unwind, and this is mediated by helicase enzymes.
The leading strand is synthesized continuously but the opposite strand is copied in short bursts of about 1000 bases, as the lagging strand template becomes available. The resulting short strands are called Okazaki fragments (after their discoverers, Reiji and Tsuneko Okazaki). Bacteria have at least three distinct DNA polymerases: Pol I, Pol II and Pol III; it is Pol III that is largely involved in chain elongation. Strangely, DNA polymerases cannot initiate DNA synthesis, but require a short primer with a free 3'-hydroxyl group. This is produced in the lagging strand by an RNA polymerase (called DNA primase) that is able to use the DNA template and synthesize a short piece of RNA around 20 bases in length. Pol III can then take over, but it eventually encounters one of the previously synthesized short RNA fragments in its path. At this point Pol I takes over, using its 5'- to 3'-exonuclease activity to digest the RNA and fill the gap with DNA until it reaches a continuous stretch of DNA. This leaves a gap between the 3'-end of the newly synthesized DNA and the 5'-end of the DNA previously synthesized by Pol III. The gap is filled by DNA ligase, an enzyme that makes a covalent bond between a 5'-phosphate and a 3'-hydroxyl group. The initiation of DNA replication at the leading strand is more complex and is discussed in detail in more specialized texts.
The new DNA strands are formed, with one strand of the parent DNA and the other is newly synthesized, this process is called semiconservative DNA replication.
DNA replication is not perfect. Errors occur in DNA replication, when the incorrect base is incorporated into the growing DNA strand. This leads to mismatched base pairs, or mispairs. DNA polymerases have proofreading activity, and a DNA repair enzymes have evolved to correct these mistakes. Occasionally, mispairs survive and are incorporated into the genome in the next round of replication. These mutations may have no consequence, they may result in the death of the organism, they may result in a genetic disease or cancer; or they may give the organism a competitive advantage over its neighbours, which leads to evolution by natural selection.
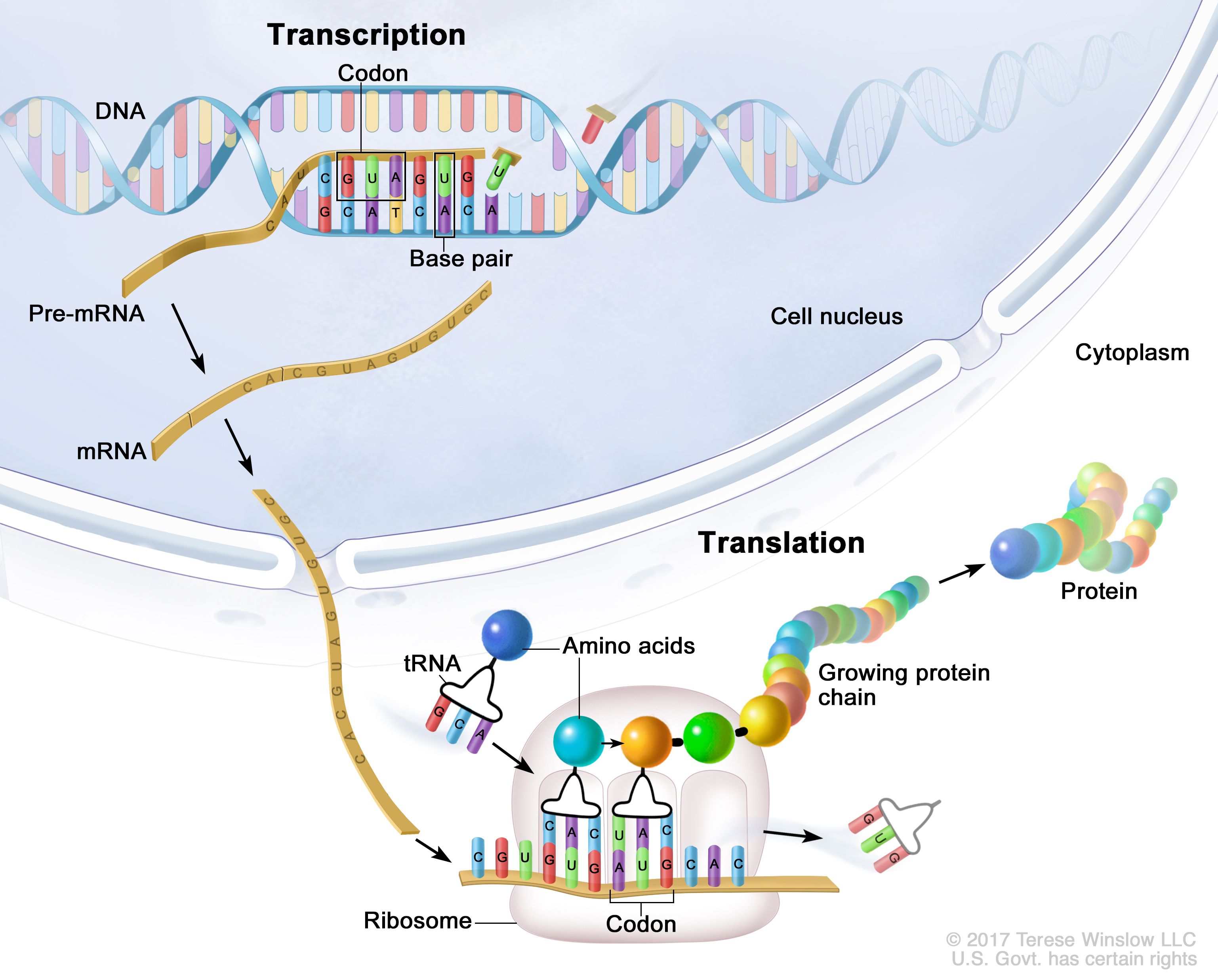
Transcription
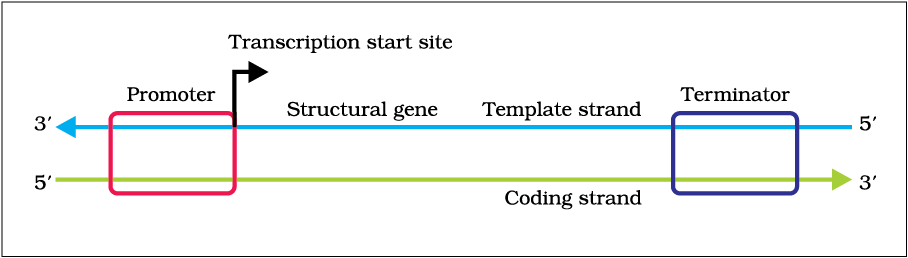
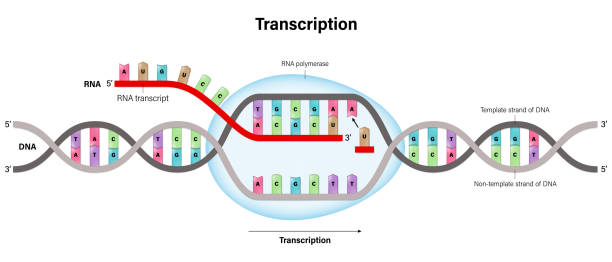
Transcription is the process by which the information is transferred from one strand of the DNA to RNA by the enzyme RNA Polymerase. The DNA strand which undergoes this process consists of three parts namely promoter, structural gene, and a terminator.
The DNA strand that synthesizes the RNA is called the template strand and the other strand is called the coding strand. The DNA-dependent RNA polymerase binds to the promoter and catalyzes the polymerization in the 3′ to 5′ direction.
Ribonucleoside triphosphates (NTPs) align along the template DNA strand, with Watson-Crick base pairing (A pairs with U). RNA polymerase joins the ribonucleotides together to form a pre-messenger RNA molecule that is complementary to a region of the template DNA strand. Transcription ends when the RNA polymerase enzyme reaches a triplet of bases that is read as a "stop" signal. The DNA molecule re-winds to re-form the double helix.
Post-Transcriptional Modifications (in Eukaryotes)
After transcription, the primary RNA transcript undergoes several modifications before it becomes mature mRNA that can be translated into a protein. These modifications include:
- 5' Capping: A modified guanine nucleotide is added to the 5' end of the RNA transcript. This cap protects the RNA from degradation and helps in the initiation of translation.
- Polyadenylation: A tail of adenine nucleotides (poly-A tail) is added to the 3' end of the RNA transcript. This tail also protects the RNA from degradation and aids in the export of the mRNA from the nucleus to the cytoplasm.
- Splicing: In this process, the introns or the non-coding sequences present in the pre-mRNA strand are spliced out such that only the exons or the coding sequence remain.
Translation
Transfer RNA
Transfer RNA adopts a well defined tertiary structure which is normally represented in two dimensions as a cloverleaf shape, Each amino acid has its own special tRNA (or set of tRNAs). For example, the tRNA for phenylalanine (tRNAPhe) is different from that for histidine (tRNAHis). Each amino acid is attached to its tRNA through the 3'-OH group.
The Process of Translation
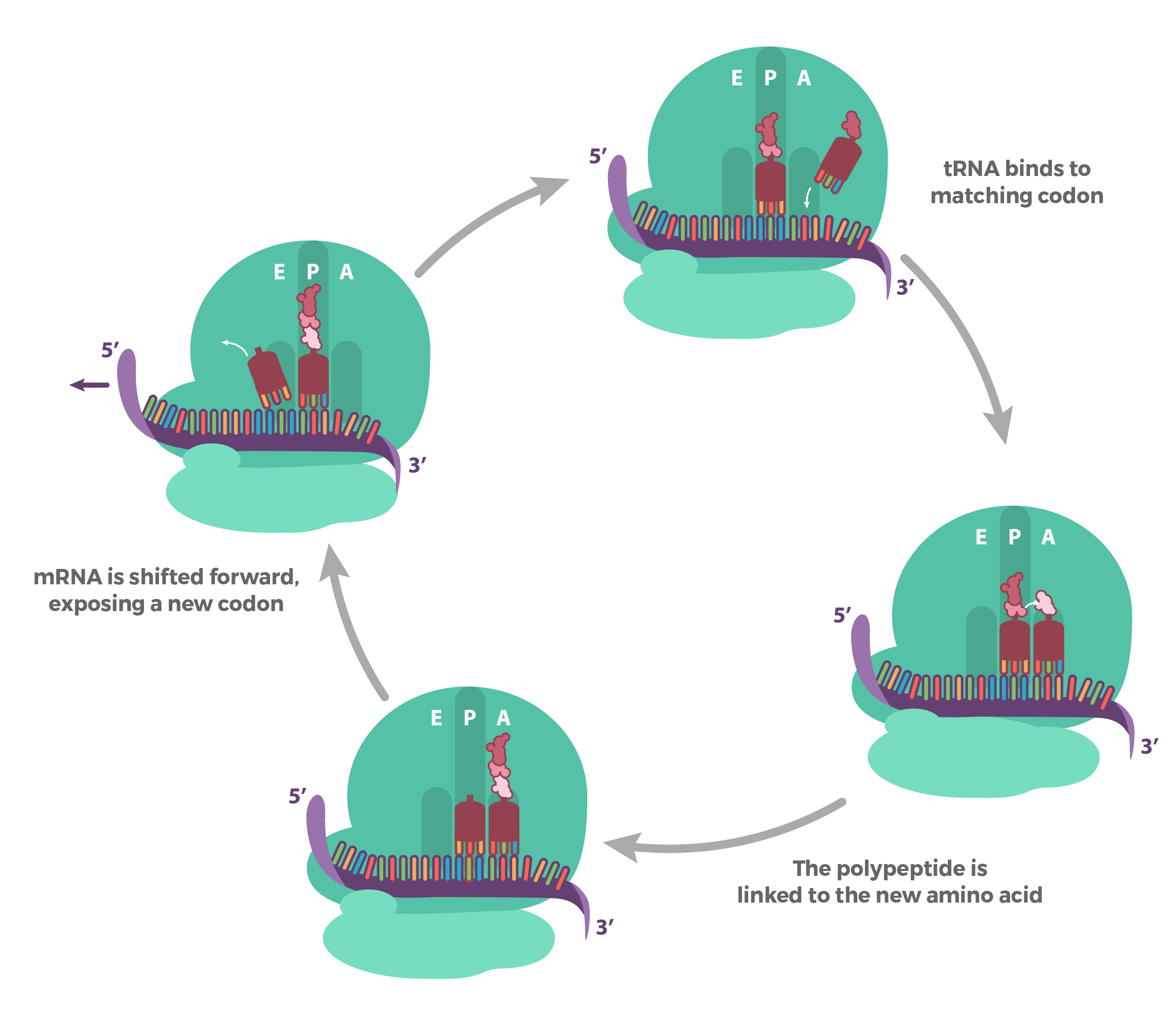
Translation is the process by which the genetic code carried by mRNA (messenger RNA) is used to build proteins, which are the molecules that do most of the work in our cells. Here's a simple breakdown of the steps involved in translation:
1. mRNA Leaves the Nucleus: After being made in the nucleus, the mRNA travels to the ribosome, the cell's protein-making machine, which is located in the cytoplasm.
2. Ribosome Binding: The ribosome attaches to the mRNA at a specific spot called the start codon. This is like finding the beginning of a recipe.
3. Reading the Code: The ribosome reads the mRNA three nucleotides at a time. Each group of three nucleotides is called a codon, and each codon codes for a specific amino acid (the building blocks of proteins).
4. tRNA Brings Amino Acids: Transfer RNA (tRNA) molecules carry amino acids to the ribosome. Each tRNA has an anticodon that matches a codon on the mRNA. When the tRNA's anticodon matches the mRNA's codon, the ribosome adds the amino acid it carries to the growing protein chain.
5. Building the Protein: The ribosome moves along the mRNA, reading codons and adding the corresponding amino acids one by one. This process continues until the ribosome reaches a stop codon, which signals the end of the protein-making process.
6. Protein Release: Once the stop codon is reached, the newly made protein is released from the ribosome and can start folding into its final shape and begin its work in the cell.
Translation is like reading a recipe (the mRNA) to make a dish (the protein), with the ribosome acting as the chef and tRNA molecules bringing the ingredients (amino acids).
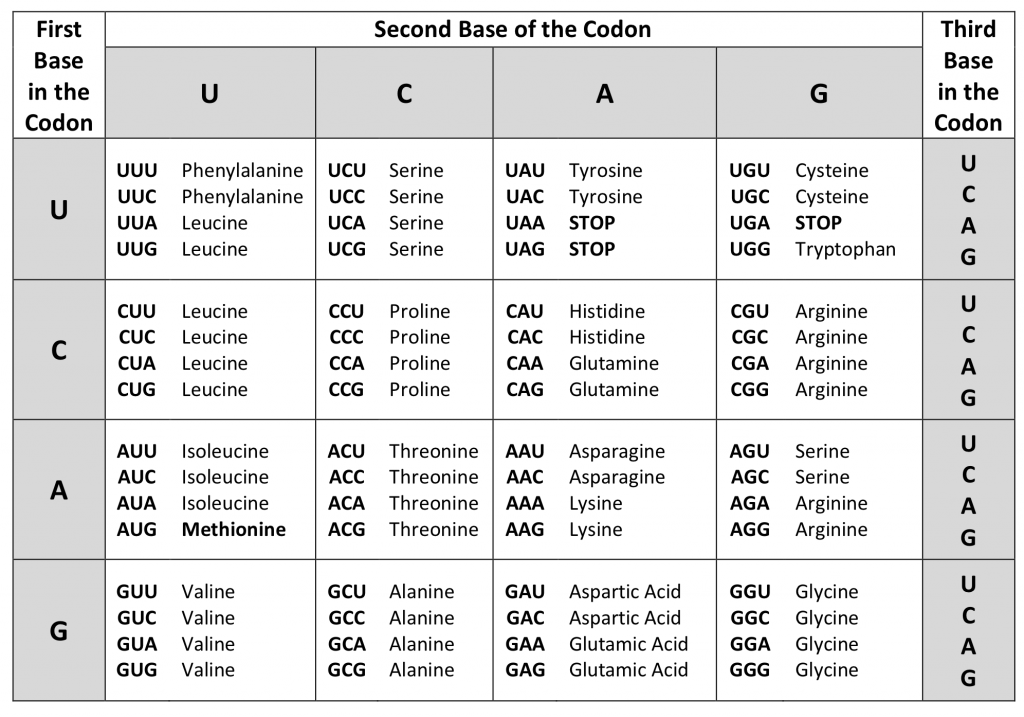
Genetic Code
Genetic code contains the information of the protein manufactured from RNA. There are basically three nucleotides and four nitrogenous bases, which collectively form a triplet codon that codes for one amino acid. Therefore, the number of possible amino acids range to 4 x 4 x 4 = 64 amino acids. There are 20 naturally existing amino acids.
The genetic code degenerates. This was explained by the features of the genetic code, according to which a few amino acids are coded by more than one codon thus causing them to degenerate. Each codon codes for only one specific amino acid and the codes are universal irrespective of the type of organism. Out of the 64 codons, 3 are stop codons which stop the process of transcription and one of the codons is an initiator codon i.e. AUG coding for Methionine.
Exceptions to the central dogma of molecular biology
Although the dogma has been widely accepted since its formulation in the 1950s, some recent discoveries have cast doubt on its rigidity. For example, it has been shown that some viruses can synthesize proteins from RNA without going through DNA, and that certain types of RNA can have functions beyond serving as intermediates in protein synthesis (such as interference RNA).
Another exception is retrotranscription, the process by which RNA is used as a template to synthesize DNA. This process is carried out by the reverse transcriptase enzyme, and is present in retroviruses, such as HIV.
A video on Central Dogma
Sources:
Image Credits:



Comments
Post a Comment